ACCRE Introduction
Access this presentation

Do you have these situations?
Do you have a 8G/16G laptop and screaming at it/yourself when you try to load a imaging/EHR/Genomic data?
Do you max out your laptop’s fan doing simulation during class and all the people are looking at you (Yeji)
Or do you have 100,000,000,000 simulations need to run and so afraid to close your laptop/want to speed it up?
Or you just want to be cool
ACCRE!!
It’s a cluster, with a lot of CPU and even more RAM than you need
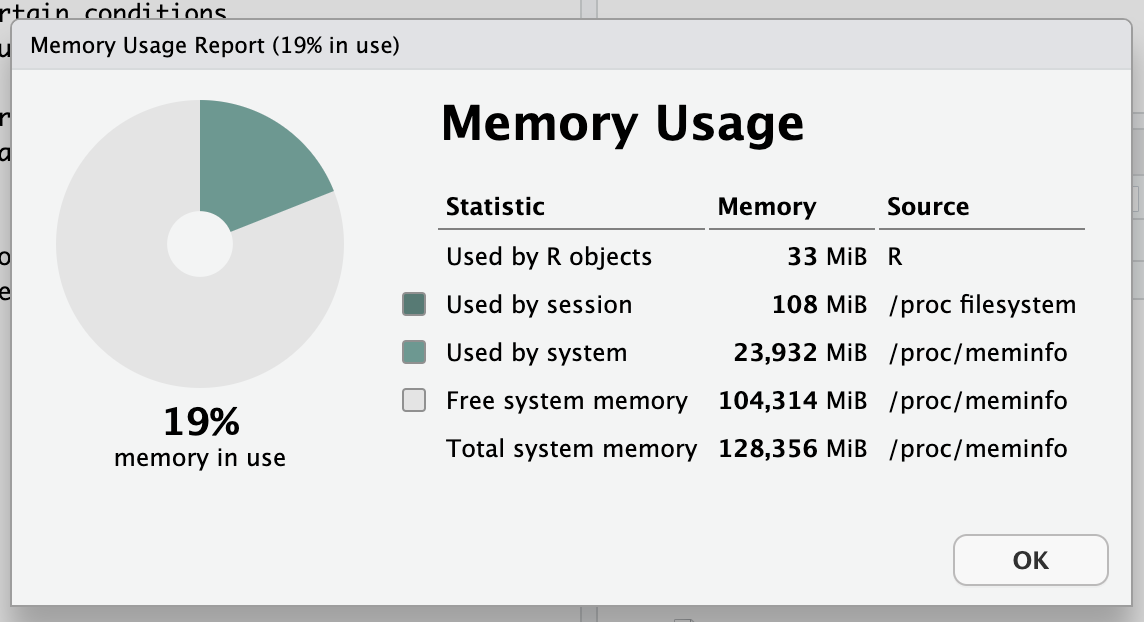
128G Virtual Machine
Why people not using ACCRE
I don’t know how to use Linux
This is Linux
I promise you you will not see a single line of linux code for the first 1/3 of this tutorial
Visual Portal

File
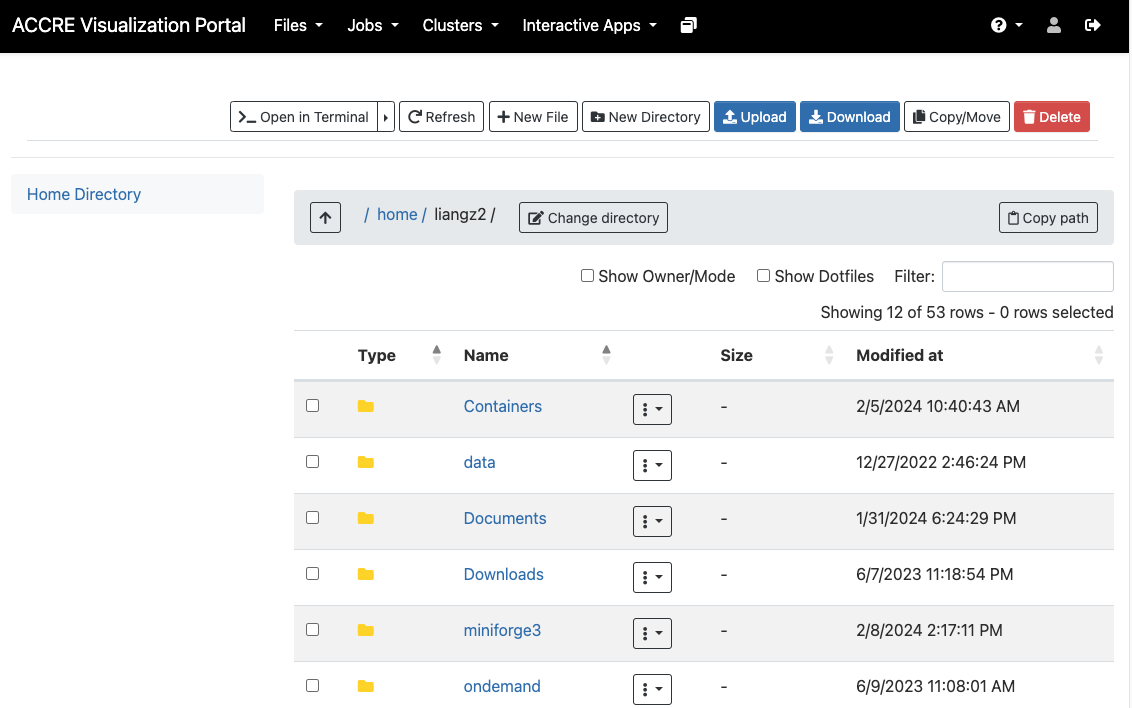
Interactive Apps
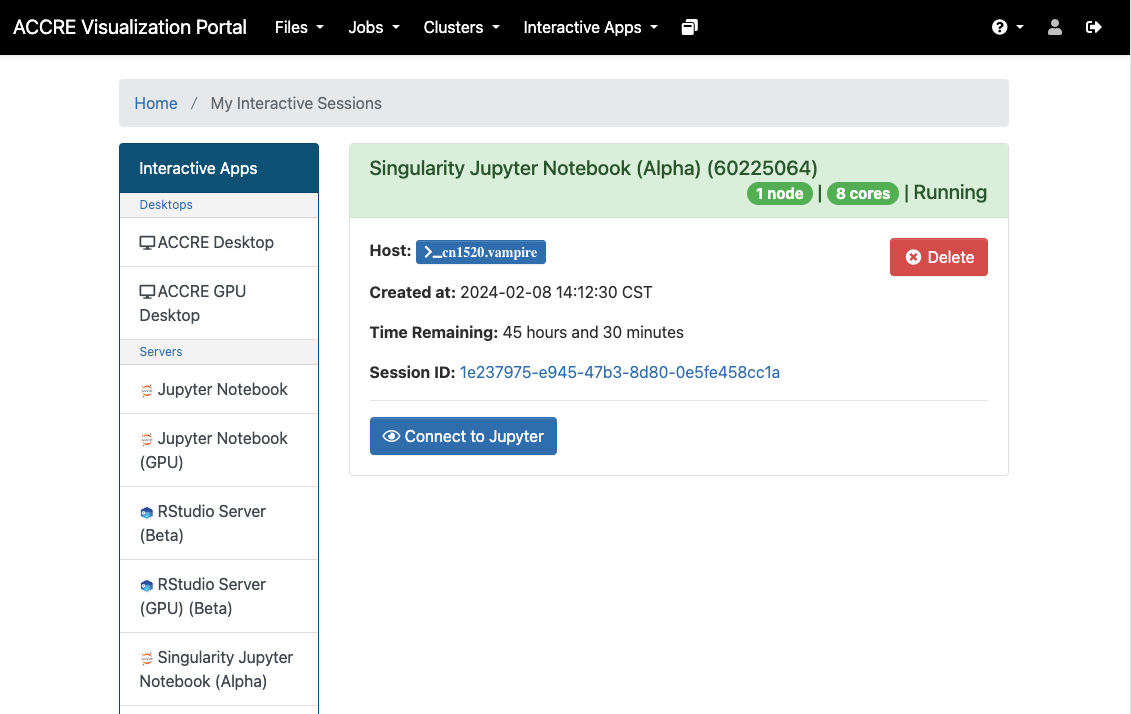
Interactive Apps

Rstudio Server and Jupyter are your friends and most efficient workspace in ACCRE
Don’t even try the desktop it’s horrible
See it’s not that bad
- With Rstudio and Jupyter Notebook you can gain access to the RAM you need for your code-writing
- No a single line of Linux code so far
- But it doesn’t necessarily speed up your work
Background Job
This is the most powerful tools of ACCRE
Run code, go to sleep
- How to use SCREEN
- How to use slurm
- How to use slurm to run 1000 simulation simultaneously
Screen
There’s a very simple tutorial here
After installed screen
Slurm
A tutorial is here
Slurm allows you to run as much simulations as you want in the background, simultaneously*
All you need is
- You simulation code(*.R,*.python)
- A slurm file(Don’t worry, they have templates there)
Slurm
Have your simulation code ready(*.R, *.python)
Slurm
Create a file like simulation.slurm
#!/bin/bash
#SBATCH --mail-user=vunetid@vanderbilt.edu
#SBATCH --mail-type=ALL
#SBATCH --ntasks=1
#SBATCH --time=00:05:00
#SBATCH --mem=250M
#SBATCH --array=0-24
#SBATCH --output=wdi-by-year-%a.out
module load GCC OpenMPI R
R --version
echo "SLURM_JOBID: " $SLURM_JOBID
echo "SLURM_ARRAY_TASK_ID: " $SLURM_ARRAY_TASK_ID
echo "SLURM_ARRAY_JOB_ID: " $SLURM_ARRAY_JOB_ID
Rscript wdi-by-year.r $SLURM_ARRAY_TASK_IDSlurm
Then submit your job in terminal
Parallel Job!!!
This is how ACCRE going to boost your research to meet the deadline every Friday
Same for slurm,
There is a tutorial of how to do Parallel computing in ACCRE
- Using ACCRE doesn’t necessarily speed up your job if you have like 500 3-second simulations
- Resources are shared among all of us, so sometime all of us are using and has waiting line for the servers
Environments
If you are afraid of Linux, more like you are afraid of building the environment
In ACCRE, environment was built and load with
module. It is easy to use, but most of the time it is out-of-date(R is 4.0.5)conda/mambais the easiest way to build a custom environmentDocker container is my recommended way to build environment
Links for installation were attached
Module & Mamba
- Using Module
Docker
This has a little bit of learning curve, but once you know how to do it, you. can do this:
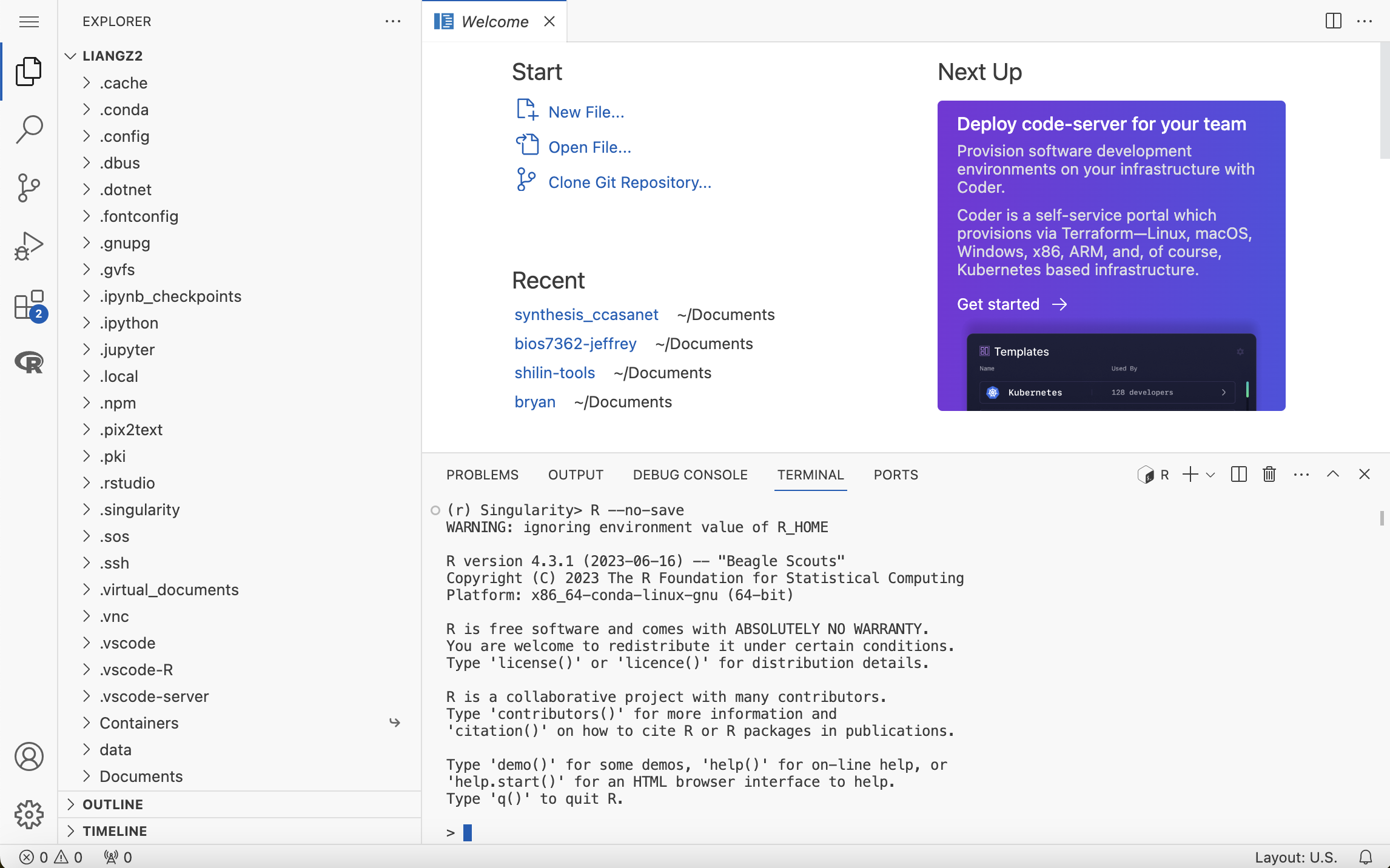
A tutorial of Singularity(Accre’s Docker) is here
Docker
And launch it with the interaction-app
Docker
Build your own container:
What I did here is install proxy for vs-code in jupyter, so that I can use vs-code in ACCRE
FROM ubuntu:22.04 #core
#start building environment
RUN apt-get update && apt-get install -y \
wget \
curl \
git \
&& rm -rf /var/lib/apt/lists/*
RUN curl -fsSL https://code-server.dev/install.sh | sh
RUN code-server --install-extension ms-python.python && \
code-server --install-extension ms-toolsai.jupyter
WORKDIR /app
RUN wget "https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh" -O miniconda.sh && \
bash miniconda.sh -b -p /app/conda -f && \
rm miniconda.sh
ENV PATH=$PATH:/app/conda/bin
ARG PATH=$PATH:/app/conda/bin
RUN mamba install -y\
jupyterlab \
notebook \
jupyter-vscode-proxy && \
mkdir /app/.jupyter
EXPOSE 8888 8787 8080
CMD ["jupyter-lab","--ip=0.0.0.0","--port=8888","--no-browser","--allow-root","--NotebookApp.token=''", "--NotebookApp.password=''"]Summary
ACCRE itself was built to be used with minimal knowledge of Linux
But you can expand what you can do with ACCRE with a little bit of exploring the linux world
I am sure that someone in this room has better idea to maximizing efficient from ACCRE
REMEMBER: We shared the resources so use it with others people in mind
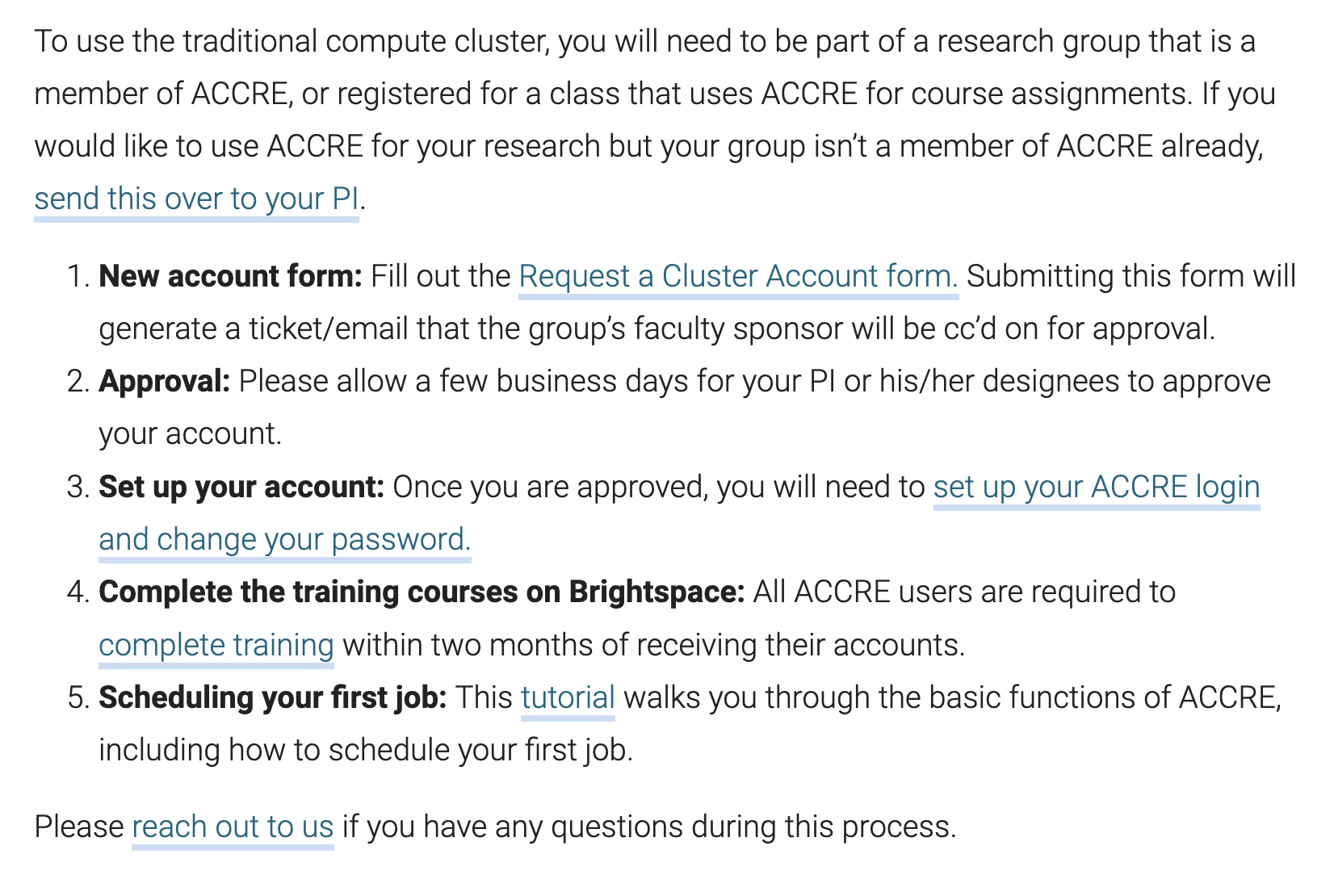
So go ahead and apply for an account in
accre.vanderbilt.edu. It comes with a training module that takes less than 1hr and very helpful.
Summary
- So go ahead and apply for an account in
accre.vanderbilt.edu. It comes with a training module that takes less than 1hr and very helpful.
Thank You
